 |
| 3D Modeling of the Human Immunodeficiency Virus [1] |
Growing up in the 1990’s and being the son of an infectious disease physician, I had always been warned of the dangers of HIV. Particularly, I was taught about the fact that there is no cure once one is infected with the virus. As I grew up and became more educated in the biological sciences, I became curious to why we essentially are able to prevent certain viral infections (such as the chicken pox virus) and not have any way to prevent other viral infections such as HIV. “Aren’t all viruses the same and why can’t we ever truly be cured of a viral infection?” The short answer, which I would come to learn, is that there is a large amount of diversity among the different viruses that can infect our bodies and hijack our essential life systems. However, a universal key mechanism of viral infections that makes them extremely hard to cure is that there is incorporation of viral genetic material into our own genome of an affected cell; essentially making the virus a part of us. Click the image below to explore the replication (life cycle) of the HIV virus.
 |
| HIV Replication Cycle [2] |
 |
| My Dad and I at dinner |
So, when I came across the article titled Experimental vaccine helps protect monkeys against AIDS-like infection on CNN Health a month back I was immediately intrigued. The article stated that the developed vaccine had been successful in treating SIV (Simian Immunodeficiency Virus), a close relative of the human HIV virus and the primary animal model virus for HIV. Although the vaccine had been successful in initially preventing clinical SIV infection in the tested monkeys, each individual monkey eventually developed SIV after repeated exposures. Despite the vaccines eventual failure to completely ameliorate the potential for SIV infection, the scientific community believes that they are now a step closer to finding a preventative cure for HIV. After reading the article and realizing that HIV has not been cured, I became curious about what kinds of treatments there are on the market to treat someone with HIV/AIDS.
I decided to preform a basic google search for “HIV/AIDS Treatments.” The search resulted with the the Mayo Clinic website article outlining the different treatments. The article stated that there are a wide variety of drug treatment options. These treatments included Non-nucleoside reverse transcriptase inhibitors (NNRTIs), nucleoside reverse transcriptase inhibitors (NRTIs), protease inhibitors (PIs), entry of fusion inhibitors, and integrase inhibitors. With many kinds of treatments to chose from, I discovered that treatment in the clinic for HIV/AIDS involves a cocktail of different treatment types. Fascinated by HIV’s ability to convert its RNA genetic code into DNA, I dug further to investigate how the different drugs on the market affect reverse transcriptase, the enzyme that is responsible to convert RNA to DNA.
The two classes of drugs that affect reverse transcriptase action are NNRTIs and NRTIs. NNRTIs act by antagonistically binding to the enzyme and halting its function where NRTIs utilize faulty versions of the building blocks that reverse transcriptase uses to convert RNA to DNA. This essentially prevents a good DNA copy of the viral genome to be produced. A key difference between NRTIs and NNRTIs is that one is a competitive inhibitor while the other is a non-competitive inhibitor of the enzymatic process. NRTI’s are competitive inhibitors. They compete with the cells normal nucleosides (DNA building blocks) for integration into the viral DNA strand. On the other hand NNRTIs are non-competitive inhibitors meaning that they are a particular kind of molecule that reduces enzyme activity by binding to a site that is different than the active site of a molecule where it does not face binding competition [3].
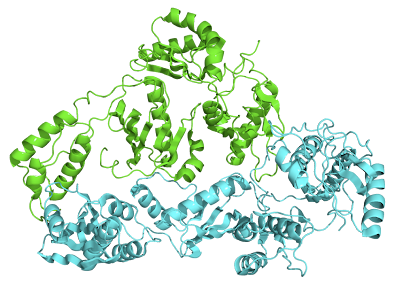 |
| Crystallographic Structure of Reverse Transcriptase where the P51 subunit is colored green and the P66 subunit is colored cyan [4] |
After another quick Goggle search for NNRTIs, I came to learn that the FDA had recently approved a new drug called Rilpivirine (trade name Edurant) in May 2011. I became instantly curious to learn the novel mechanism of action by which this drug inhibits reverse transcriptase. Like all NNRTI’s, Rilpvirine (see Marvin Sketch Below) inhibits reverse transcriptase by allosteric binding. This type of inhibition works by non-competitively binding to reverse transcriptase at a location other than the enzyme’s active site, resulting in inhibition of its function. For Reverse Transcriptase this binding site is known as the Non-Nucleoside Inhibitor Binding Pocket (NNIBP). But what about this drug makes it unique?
| Figure 2. Mouseover sends to MarvinSketch. |
Before I searched for the answer to this question, I sought to understand the structure of reverse transcriptase, specifically the NNIBP, as well as the characteristics that make a good HIV/AIDS drug. A preformed a quick Google search resulted for "reverse transcriptase" and ended up finding ample information. The first link that I clicked on was the Wikipedia page for Reverse transcriptase. From the wiki page I learned that Reverse transcriptase is a type of polymerase that is RNA-dependent DNA synthesizing and that it is made up of two distinct components, the p51 and p66 subunits. The p66 subunit contains the fingers, palm and thumb of the enzyme and the p51 subunit contains an additional set of fingers. The analogy of reverse transcriptase as a hand is used since it describes which parts of the enzyme are squeezing together to connect the different nucleosides in a chain. Use the Jmol applet below to explore the structure of Reverse Transcriptase.
The Non-nucleoside inhibitor-binding pocket (NNIBP) is located on the P66 subunit. This pocket has a characteristic positive charge and hydrophobicity due to the presence of particular amino acids including Val 106, Tyr 188, Trp 229, Leu 234, Leu 100, Phe 227, Tyr 319, Pro 236, Tyr 181, Lys 103, Gly 190. During protein synthesis these amino acids come together to form a pocket (via hydrophobic interactions) to shield themselves from the highly polar hydrophilic intracellular space. The NNIBP is located approximately 10 angstroms away from reverse transcriptase’s active binding site. The active binding site is the location where reverse transcriptase converts the viral RNA into DNA. NNRTIs target the NNIBP and causing a change in steric interactions that results in inactivation of reverse transcriptase’s function.
I found the answer to my second question "what makes a good anti-HIV/AIDs medication?" in an article published in the Journal of Medicinal Chemistry. The article stated that a good anti-HIV drug should have the following properties: (1) it should be highly active against wild-type and mutant types of HIV without allowing breakthrough, (2) it should have high oral bioavailability and long elimination half-life, allowing for once-daily oral treatment at low doses, (3) have minimal adverse effects, and (4) be easy to synthesize and formulate [5]. With a brief and basic understanding of the structure and properties of Reverse transcriptase and its inhibition binding pocket as well as the key properties of a well engineered anti-HIV drug, I was able to move forward and learn more about Rilpivirine. Specifically, how it acts as a NNRTI as well as how it is different from other NNRTIs that are available on the market.
I found the answer to my second question "what makes a good anti-HIV/AIDs medication?" in an article published in the Journal of Medicinal Chemistry. The article stated that a good anti-HIV drug should have the following properties: (1) it should be highly active against wild-type and mutant types of HIV without allowing breakthrough, (2) it should have high oral bioavailability and long elimination half-life, allowing for once-daily oral treatment at low doses, (3) have minimal adverse effects, and (4) be easy to synthesize and formulate [5]. With a brief and basic understanding of the structure and properties of Reverse transcriptase and its inhibition binding pocket as well as the key properties of a well engineered anti-HIV drug, I was able to move forward and learn more about Rilpivirine. Specifically, how it acts as a NNRTI as well as how it is different from other NNRTIs that are available on the market.
Rilpivirine is a diarylpyrimidine and has the structural formula 4-[[4-[[4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]2-pyrimidinyl]amino]benzonitrile. In lay terms, Rilpivirine is made up of three cyclic rings, two nitrile groups, and a conjugated double bond system. Like most diarylpyrimidine non-nucleoside reverse transcriptase inhibitors, I learned that Rilpivirine has characteristic flexibility that gives it the ability to maintain its activity in situations where wild-type reverse transcriptase is not present. Also, Rilpivirine’s targets certain amino acids which display low levels of mutability which helps give the drug very low resistance against mutant reverse transcriptase’s. Explore Rilpivirine's structure using the Jmol applet below.
It is Rilpivirine’s ability to interact with both mutant and wild type Reverse Transcriptase that makes it a novel NNRTI. Originally, NNRTI’s were not effective in treating patients when mutant Reverse Transcriptase was present. The resistance towards older NNRTI's was due impart to decreased torsion and flexibility among the drugs chemical bonds. Another key feature of early NNRTIs was their dependence on π -stacking interaction with specific Tyrosine groups (Y181 and Y188). Evidently, it turned out that these specific Tyrosine groups exhibit high mutation rates and accounted for the most common type of observed mutant reverse transcriptase in the clinic. Since the binding mechanism of these NNRTI’s relied on highly stable and ridged π-stacking interactions, when these amino acid groups mutated, the drugs had no way of conforming to fit the new mutant NNIBP. Rilpivirne, on the other hand, relies on a slightly different mechanism of binding and employs greater torisonal flexibility. The torsion angle diagram shows the degree of rotation around certain sigma bonds when Rilpivirine is bound to its binding pocket. Despite the single degree of rotational difference between the wild-type and mutant version of Reverse Transcriptase, researchers believe it is this extra degree that makes all the difference.
 |
| Rilpivirine Torsion Degrees in Wild-Type & Mutant Reverse Transcriptase [6] |
Instead of targeting Y181 and Y188, Rilpivirine targets a specific Tryptophan group (W299). In Reverse Transcriptase mutant analysis, it has been shown that this specific Tryptophan group shows stability, with regards to mutability, across all mutants making it the perfect target for a NNRTI with low levels of drug resistance. As shown in the figure below, Rilpivirine’s nitrile groups give it the ability to specifically target and interact with Tryptophan 229 and Tyrosine 318. These highly polar functional groups are able to interact via dipole-dipole interactions with W229 and Y318 helping to Rilpivirine to settle into the binding pocket. With regards to W229, the nitrile group interacts via an edge to face interaction. It is also possible that these nitrile groups could be undergoing H-bonding even though they are poor hydrogen acceptors. Like previous NNRTIs, Rilpivirine relies on π-stacking, hydrophobic interactions as well as electron rich and electron deficient interactions.
 |
| Rilpivirine in the NNIBP [5] |
Although mechanistically the theory behind how Rilpivirine would interact with Reverse Transcriptase and inhibit its function makes sense, it is often found that in vivo, drugs and treatments have very unpredictable actions. With all drug development there is the inherent risk that the drug you developed to treat a certain disease will cause harm in another part of the body. Since Rilpivirine was approved by the FDA, it is safe to say that it is a safe and effective drug to treat HIV. However, how does Rilpivirine compare to other drugs that have been proven to be successful NNRTI drug treatments. A quick search on the FDA’s website returned the clinical data from its stage three clinical drug trials in which a comparison study between Rilpivirine and the leading drug on the market, Efavirenz, was conducted. See graphs below that compare Rilpivirine and Efavirenz effectiveness by comparing measured CD4 cell counts and viral RNA concentration of treated patients.
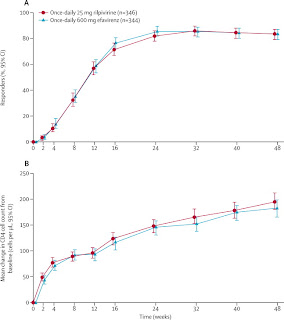 |
The data shows that Rilpivirine is an equally effective drug in comparison to Efavirenz. From the graphs above both CD4 cell count (graph B) and viral load (graph A) for patients treated with Rilpivirine showed similar disease efficacy. Over time, CD4 cell counts increased and Viral Load plateaued in treated patients. Despite the similar clinical results in patients treated with either Rilpivirine or Efavirenz, it is important to remember that Rilpivirine is overall a better drug of choice. It shows low levels of resistance to differing mutant forms of Reverse Transcriptase, has been shown to have fewer side effects while still meeting the effectiveness of previous drugs on the market. Rilpivirine in conjuction with other HIV treatments should help to improve the lives of living with this disease. Although, the cure for HIV/AIDS has not been discovered it is these small advances in medical treatment and research that help to pave the way for hope and aspiration that one day a cure of HIV will be discovered.
References:
[1] "Human Immunodeficiency Virus Model." Human Immunodeficiency Virus Model. Ed. Ivan Konstantinov. Visual Science, 8 Sept. 2010. Web. 12 May 2012 <http://visualscience.ru/en/projects/hiv/illustrations/>.
[2] National Institute of Allergy and Infectious Disease. "HIV Replication Cycle." HIV Replication Cycle. National Institute of Health, 3 Apr. 2012. Web. 12 May 2012. <http://www.niaid.nih.gov/topics/HIVAIDS/Understanding/Biology/pages/hivreplicationcycle.aspx>.
[3] National Institues of Health Chemical Genomics Center. "Types of Inhibition." - Assay Guidance Wiki. National Institues of Health Chemical Genomics Center, 20 Sept. 2010. Web. 12 May 2012. <http://assay.nih.gov/assay/index.php/Types_of_Inhibition>.
[4] "RCSB Protein Data Bank - RCSB PDB - 1HMV Structure Summary." RCSB Protein Data Bank - RCSB PDB - 1HMV Structure Summary. RCSB Protein Data Bank. Web. 12 May 2012. <http://www.rcsb.org/pdb/explore/explore.do?structureId=1hmv>.
[5] Janssen, Paul A. J., Paul J. Lewi, Eddy Arnold, Frits Daeyaert, Marc De Jonge, Jan Heeres, Luc Koymans, Maarten Vinkers, Jérôme Guillemont, Elisabeth Pasquier, Mike Kukla, Don Ludovici, Koen Andries, Marie-Pierre De Béthune, Rudi Pauwels, Kalyan Das, Art D. Clark,, Yulia Volovik Frenkel, Stephen H. Hughes, Bart Medaer, Fons De Knaep, Hilde Bohets, Fred De Clerck, Ann Lampo, Peter Williams, and Paul Stoffels. "In Search of a Novel Anti-HIV Drug: Multidisciplinary Coordination in the Discovery of 4-[[4-[[4-[(1)-2-Cyanoethenyl]-2,6-dimethylphenyl]amino]-2- Pyrimidinyl]amino]benzonitrile (R278474, Rilpivirine)." Journal of Medicinal Chemistry 48.6 (2005): 1901-909. Print.
[6] Lansdon, Eric B., Katherine M. Brendza, Magdeleine Hung, Ruth Wang, Susmith Mukund, Debi Jin, Gabriel Birkus, Nilima Kutty, and Xiaohong Liu. "Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design." Journal of Medicinal Chemistry 53.10 (2010): 4295-299. Print.
[8] Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with
HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial; Dr, Prof Jean-Michel
Molina MD,Pedro Cahn MD,Beatriz Grinsztejn MD,Prof Adriano Lazzarin MD,Anthony Mills
MD,Prof Michael Saag MD,Prof Khuanchai Supparatpinyo MD,Prof Sharon Walmsley MD,Herta
Crauwels PhD,Laurence T Rimsky PhD,Simon Vanveggel MSc,Katia Boven MD,on behalf of the
ECHO study group The Lancet - 16 July 2011 ( Vol. 378, Issue 9787, Pages 238-246 ) DOI:
10.1016/S0140-6736(11)60936-7





+.jpg)
+.gif)











